

The goal of sooty is to provide access to data of relevance to Southern Ocean research.
To see what files we know about in object storage, use
library(sooty)
(files <- sooty_files())
#> # A tibble: 118,292 × 7
#> date source Dataset Bucket Key Protocol Host
#> <dttm> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 1981-09-01 00:00:00 /vsicurl/https://pro… oisst-… idea-… www.… /vsicurl http…
#> 2 1981-09-02 00:00:00 /vsicurl/https://pro… oisst-… idea-… www.… /vsicurl http…
#> 3 1981-09-03 00:00:00 /vsicurl/https://pro… oisst-… idea-… www.… /vsicurl http…
#> 4 1981-09-04 00:00:00 /vsicurl/https://pro… oisst-… idea-… www.… /vsicurl http…
#> 5 1981-09-05 00:00:00 /vsicurl/https://pro… oisst-… idea-… www.… /vsicurl http…
#> 6 1981-09-06 00:00:00 /vsicurl/https://pro… oisst-… idea-… www.… /vsicurl http…
#> 7 1981-09-07 00:00:00 /vsicurl/https://pro… oisst-… idea-… www.… /vsicurl http…
#> 8 1981-09-08 00:00:00 /vsicurl/https://pro… oisst-… idea-… www.… /vsicurl http…
#> 9 1981-09-09 00:00:00 /vsicurl/https://pro… oisst-… idea-… www.… /vsicurl http…
#> 10 1981-09-10 00:00:00 /vsicurl/https://pro… oisst-… idea-… www.… /vsicurl http…
#> # ℹ 118,282 more rowsThe main columns of interest are date and
source and Dataset, the source is
a directly useable source identifier that a GDAL-enabled package can
read. (See examples below).
We can get a very simple model of a “logical dataset” by honing in
one Dataset in particular. First let’s make a summary from
everything:
library(sooty)
sooty_files() |>
dplyr::mutate(date = as.Date(date)) |> dplyr::group_by(Dataset) |>
dplyr::summarize(earliest = min(date), latest = max(date), n = dplyr::n()) |>
dplyr::arrange(Dataset, earliest)
#> # A tibble: 14 × 4
#> Dataset earliest latest n
#> <chr> <date> <date> <int>
#> 1 BREMEN-SEAICE-SMOS-north 2010-05-01 2025-05-20 5474
#> 2 BREMEN-SEAICE-SMOS-south 2010-10-01 2016-05-03 12
#> 3 NSIDC_SEAICE_PS_N25km 1978-10-26 2025-05-20 17003
#> 4 NSIDC_SEAICE_PS_S25km 1978-10-26 2025-05-20 17003
#> 5 SEALEVEL_GLO_PHY_L4 1993-01-01 2025-05-21 11715
#> 6 antarctica-amsr2-asi-s3125-tif 2012-07-02 2025-05-20 4695
#> 7 ccmp-wind-product-v2 1993-01-02 2024-08-31 11552
#> 8 esacci-oc-l3s-chlor-a-merged-5day 1997-09-03 2025-03-27 2019
#> 9 esacci-oc-l3s-chlor-a-merged-annual 1997-05-22 2024-05-22 28
#> 10 esacci-oc-l3s-chlor-a-merged-daily 1997-09-04 2025-03-31 10045
#> 11 esacci-oc-l3s-chlor-a-merged-monthly 1997-09-01 2025-03-01 331
#> 12 ghrsst-tif 2002-06-01 2025-05-20 8390
#> 13 oisst-avhrr-v02r01 1981-09-01 2025-05-20 15968
#> 14 oisst-tif 1981-09-01 2025-05-18 14057library(terra)
#> terra 1.8.50
file <- files$source[which.max(files$date) ]
print(file)
#> [1] "/vsicurl/https://projects.pawsey.org.au/idea-sealevel-glo-phy-l4-nrt-008-046/data.marine.copernicus.eu/SEALEVEL_GLO_PHY_L4_NRT_008_046/cmems_obs-sl_glo_phy-ssh_nrt_allsat-l4-duacs-0.125deg_P1D_202411/2025/05/nrt_global_allsat_phy_l4_20250521_20250521.nc"
rast(file)
#> class : SpatRaster
#> dimensions : 1440, 2880, 10 (nrow, ncol, nlyr)
#> resolution : 0.125, 0.125 (x, y)
#> extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax)
#> coord. ref. : +proj=longlat +a=6378136.3 +rf=298.257 +no_defs
#> sources : nrt_global_allsat_phy_l4_20250521_20250521.nc:sla
#> nrt_global_allsat_phy_l4_20250521_20250521.nc:err_sla
#> nrt_global_allsat_phy_l4_20250521_20250521.nc:ugosa
#> ... and 7 more sources
#> varnames : sla (Sea level anomaly)
#> err_sla (Formal mapping error)
#> ugosa (Geostrophic velocity anomalies: zonal component)
#> ...
#> names : sla, err_sla, ugosa, err_ugosa, vgosa, err_vgosa, ...
#> unit : m, m, m/s, m/s, m/s, m/s, ...
#> time (days) : 2025-05-21The available datasets can be found from the table.
dplyr::distinct(sooty_files(), Dataset)
#> # A tibble: 14 × 1
#> Dataset
#> <chr>
#> 1 oisst-avhrr-v02r01
#> 2 SEALEVEL_GLO_PHY_L4
#> 3 NSIDC_SEAICE_PS_S25km
#> 4 NSIDC_SEAICE_PS_N25km
#> 5 antarctica-amsr2-asi-s3125-tif
#> 6 ghrsst-tif
#> 7 ccmp-wind-product-v2
#> 8 BREMEN-SEAICE-SMOS-south
#> 9 BREMEN-SEAICE-SMOS-north
#> 10 oisst-tif
#> 11 esacci-oc-l3s-chlor-a-merged-monthly
#> 12 esacci-oc-l3s-chlor-a-merged-daily
#> 13 esacci-oc-l3s-chlor-a-merged-5day
#> 14 esacci-oc-l3s-chlor-a-merged-annualThere are data sets like the 25km resolution south polar stereographic sea ice concentration, available via the dataset identifier `:
icefiles <- sooty_files() |> dplyr::filter(Dataset == "NSIDC_SEAICE_PS_S25km")
dplyr::glimpse(icefiles)
#> Rows: 17,003
#> Columns: 7
#> $ date <dttm> 1978-10-26, 1978-10-27, 1978-10-28, 1978-10-29, 1978-10-30, …
#> $ source <chr> "/vsicurl/https://projects.pawsey.org.au/idea-10.5067-mpyg15w…
#> $ Dataset <chr> "NSIDC_SEAICE_PS_S25km", "NSIDC_SEAICE_PS_S25km", "NSIDC_SEAI…
#> $ Bucket <chr> "idea-10.5067-mpyg15waa4wx", "idea-10.5067-mpyg15waa4wx", "id…
#> $ Key <chr> "n5eil01u.ecs.nsidc.org/PM/NSIDC-0051.002/1978.10.26/NSIDC005…
#> $ Protocol <chr> "/vsicurl", "/vsicurl", "/vsicurl", "/vsicurl", "/vsicurl", "…
#> $ Host <chr> "https://projects.pawsey.org.au", "https://projects.pawsey.or…You can immediately see what range of dates is available:
range(icefiles$date)
#> [1] "1978-10-26 UTC" "2025-05-20 UTC"And, explore exactly what source files are available:
range(diff(icefiles$date)) ## there are some gaps, it's every two days to start and some are missing
#> Time differences in days
#> [1] 1 7
diff(range(icefiles$date)) ## the number of potential data days
#> Time difference of 17008 days
nrow(icefiles) ## the actual number of data days
#> [1] 17003This is a very experimental begin at replacing raadtools with a package anyone can use.
We can read from the datasets with a GDAL-ready package, such as terra. But note that we need to set a configuration first, and we need to unset it after, this is WIP.
amsrfiles <- dataset("antarctica-amsr2-asi-s3125-tif")@source
#> Warning in dataset("antarctica-amsr2-asi-s3125-tif"): 'dataset' is deprecated.
#> Use 'datasource' instead.
#> See help("Deprecated")
library(terra)
(r <- rast(tail(amsrfiles$source, 1)))
#> class : SpatRaster
#> dimensions : 2656, 2528, 1 (nrow, ncol, nlyr)
#> resolution : 3125, 3125 (x, y)
#> extent : -3950000, 3950000, -3950000, 4350000 (xmin, xmax, ymin, ymax)
#> coord. ref. : WGS 84 / NSIDC Sea Ice Polar Stereographic South (EPSG:3976)
#> source : asi-AMSR2-s3125-20250520-v5.4.tif
#> color table : 1
#> name : asi-AMSR2-s3125-20250520-v5.4
r[r > 100] <- NA
plot(r[[nlyr(r)]] * 1, main = format(max(icefiles$date)))
ghrsst <- dataset("ghrsst-tif")
#> Warning in dataset("ghrsst-tif"): 'dataset' is deprecated.
#> Use 'datasource' instead.
#> See help("Deprecated")
sstfile <- ghrsst@source$source[match(max(icefiles$date), ghrsst@source$date)]
sst <- rast(sprintf("vrt://%s?ovr=4", sstfile))
ct <- as.contour(crop(sst, ext(-180, 180, -90, -40)))
plot(project(ct, crs(r)), add = TRUE, col = "hotpink")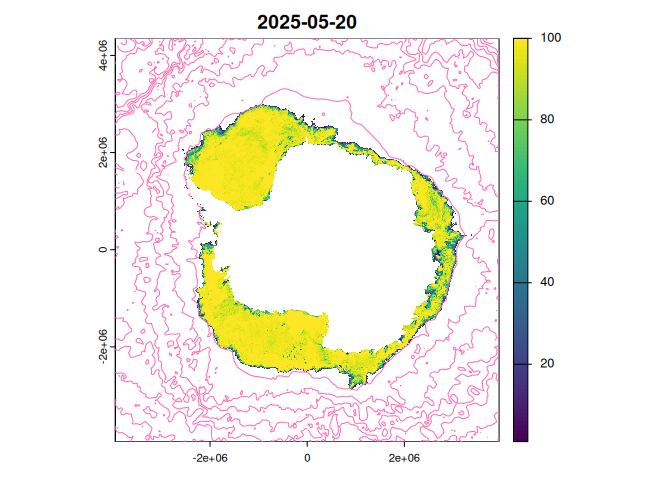
That should be the state of the sea ice in the Southern Ocean at the latest available date, sea ice concentration from passive microwave at 3.125km resolution, by the AWI artist sea ice group.
sooty_files()Dataset is the main grouping value, and files from
different Datasets are otherwise non-relatable, be it by
file format, available variable/s, grid specification (spatial extent
and resolution), or temporal organization. We guarantee that within a
Dataset, the files are ordered and unique by date(-time) and that they
are exactly relatable spatially and (mostly, or usually also) by
variable/s. The time series may not be regular or complete, but usually
is.
We retain the component parts of source, as
Bucket, Key, Protocol, and
Host as this is object storage and these are the separable
parts of addressing objects in different ways, and we’ve reserved that
for future usage. (We use https:// protocol by default to
avoid any required configurations). Get in touch if you have any
questions about this.
We have an in-progress class for dataset handling. Create a ‘dataset()’ and set an id.
ds <- dataset()
#> Warning in dataset(): 'dataset' is deprecated.
#> Use 'datasource' instead.
#> See help("Deprecated")
ds@id <- "NSIDC_SEAICE_PS_S25km"
ds
#> <sooty::dataset>
#> @ id : chr "NSIDC_SEAICE_PS_S25km"
#> @ n : int 17003
#> @ mindate: POSIXct[1:1], format: "1978-10-26"
#> @ maxdate: POSIXct[1:1], format: "2025-05-20"
#> @ source : tibble [17,003 × 7] (S3: tbl_df/tbl/data.frame)
#> $ date : POSIXct[1:17003], format: "1978-10-26" "1978-10-27" ...
#> $ source : chr [1:17003] "/vsicurl/https://projects.pawsey.org.au/idea-10.5067-mpyg15waa4wx/n5eil01u.ecs.nsidc.org/PM/NSIDC-0051.002/1978"| __truncated__ "/vsicurl/https://projects.pawsey.org.au/idea-10.5067-mpyg15waa4wx/n5eil01u.ecs.nsidc.org/PM/NSIDC-0051.002/1978"| __truncated__ "/vsicurl/https://projects.pawsey.org.au/idea-10.5067-mpyg15waa4wx/n5eil01u.ecs.nsidc.org/PM/NSIDC-0051.002/1978"| __truncated__ "/vsicurl/https://projects.pawsey.org.au/idea-10.5067-mpyg15waa4wx/n5eil01u.ecs.nsidc.org/PM/NSIDC-0051.002/1978"| __truncated__ ...
#> $ Dataset : chr [1:17003] "NSIDC_SEAICE_PS_S25km" "NSIDC_SEAICE_PS_S25km" "NSIDC_SEAICE_PS_S25km" "NSIDC_SEAICE_PS_S25km" ...
#> $ Bucket : chr [1:17003] "idea-10.5067-mpyg15waa4wx" "idea-10.5067-mpyg15waa4wx" "idea-10.5067-mpyg15waa4wx" "idea-10.5067-mpyg15waa4wx" ...
#> $ Key : chr [1:17003] "n5eil01u.ecs.nsidc.org/PM/NSIDC-0051.002/1978.10.26/NSIDC0051_SEAICE_PS_S25km_19781026_v2.0.nc" "n5eil01u.ecs.nsidc.org/PM/NSIDC-0051.002/1978.10.27/NSIDC0051_SEAICE_PS_S25km_19781027_v2.0.nc" "n5eil01u.ecs.nsidc.org/PM/NSIDC-0051.002/1978.10.28/NSIDC0051_SEAICE_PS_S25km_19781028_v2.0.nc" "n5eil01u.ecs.nsidc.org/PM/NSIDC-0051.002/1978.10.29/NSIDC0051_SEAICE_PS_S25km_19781029_v2.0.nc" ...
#> $ Protocol: chr [1:17003] "/vsicurl" "/vsicurl" "/vsicurl" "/vsicurl" ...
#> $ Host : chr [1:17003] "https://projects.pawsey.org.au" "https://projects.pawsey.org.au" "https://projects.pawsey.org.au" "https://projects.pawsey.org.au" ...
ds@source
#> # A tibble: 17,003 × 7
#> date source Dataset Bucket Key Protocol Host
#> <dttm> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 1978-10-26 00:00:00 /vsicurl/https://pro… NSIDC_… idea-… n5ei… /vsicurl http…
#> 2 1978-10-27 00:00:00 /vsicurl/https://pro… NSIDC_… idea-… n5ei… /vsicurl http…
#> 3 1978-10-28 00:00:00 /vsicurl/https://pro… NSIDC_… idea-… n5ei… /vsicurl http…
#> 4 1978-10-29 00:00:00 /vsicurl/https://pro… NSIDC_… idea-… n5ei… /vsicurl http…
#> 5 1978-10-30 00:00:00 /vsicurl/https://pro… NSIDC_… idea-… n5ei… /vsicurl http…
#> 6 1978-10-31 00:00:00 /vsicurl/https://pro… NSIDC_… idea-… n5ei… /vsicurl http…
#> 7 1978-11-01 00:00:00 /vsicurl/https://pro… NSIDC_… idea-… n5ei… /vsicurl http…
#> 8 1978-11-02 00:00:00 /vsicurl/https://pro… NSIDC_… idea-… n5ei… /vsicurl http…
#> 9 1978-11-03 00:00:00 /vsicurl/https://pro… NSIDC_… idea-… n5ei… /vsicurl http…
#> 10 1978-11-04 00:00:00 /vsicurl/https://pro… NSIDC_… idea-… n5ei… /vsicurl http…
#> # ℹ 16,993 more rowsPlease note that the idt project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.